Office of Research & Development |
 |
Office of Research & Development |
 |

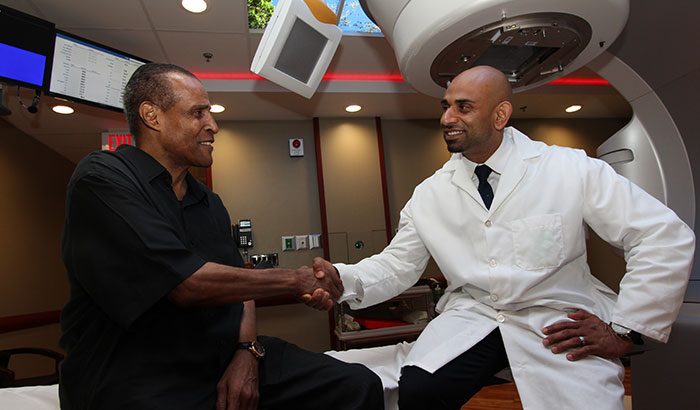
Clarence Massey (left) received treatment for his prostate cancer at the Brooklyn campus of the VA NY Harbor Healthcare System. He is seen here with radiation therapist Nader Girgis, who was part of his treatment team. (Photo by Mitch Mirkin)
May 1, 2019
By Mitch Mirkin
VA Research Communications
As an Air Force mechanic in the 1960s, Clarence Massey worked on bombers and fighter jets in Vietnam.
More than four decades later, in 2010, he became immersed in another battle. The resident of New York City’s historic Harlem neighborhood underwent nine weeks of radiation therapy for prostate cancer.
The treatment itself wasn’t too bad, he recalls, but the side effects hit him “like a ton of bricks,” says Massey, now 72.
Massey’s outcome was a good one. He gives credit to his VA doctors, but, he says, “Mostly I give credit to God. I’m blessed.”
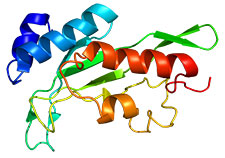
This computer-generated 3D model shows the structure of a protein encoded by tumor susceptibility gene 101. Mutations in this gene are common in breast cancer. (Image via Wikipedia)
In the years since Massey’s bout with cancer, biomedicine has come a long way—especially when it comes to analyzing genes and proteins. VA and two federal partners—the Department of Defense and the National Cancer Institute—are looking to harness the power of this science through an effort dubbed APOLLO, in the spirit of the famous space mission that landed men on the moon. The research program is described in an article now online in the journal Clinical Pharmacology & Therapeutics.
The tri-agency project launched in 2016 under the federal Cancer Moonshot, a broader effort likewise inspired by the space theme.
The full title of APOLLO sounds complex: Applied Proteogenomics Organizational Learning and Outcomes. But the overarching goal is simple: individualize cancer treatment. That’s what “precision oncology” is all about. It’s part of the personalized medicine movement. In cancer, the stakes are perhaps higher than with most other diseases.
The thinking is that lives will be saved—and plenty of nasty side effects spared—if doctors can target and kill tumors like smart bombs destroy enemy bunkers, using very specific information about the patients themselves, and about the cancers growing within them.
"Each patient is unique, and each tumor is unique."
“Each patient is unique, and each tumor is unique,” explains Dr. Craig Shriver, one of the architects of APOLLO. He directs the Murtha Cancer Center Research Program in the department of surgery at the Uniformed Services University of the Health Sciences, in Bethesda, Maryland.
APOLLO, he says, is a research project aimed at collecting and analyzing a wide array of data from cancer patients, analyzing it all with the help of sophisticated technology and scientists worldwide, and ultimately using the findings to determine the best precision treatment for a given patient. Importantly, the input will also include long-term outcomes of patients, and their responses to surveys as they progress through care.
The main beneficiaries in the short term will be Veterans and active duty troops, as cancer care at VA and military hospitals increasingly incorporates knowledge gained through APOLLO. But the knowledge will eventually filter into medical care at large, so cancer patients everywhere can see better treatment. Data-sharing resources developed by the National Cancer Institute or its partners will play a key role in that phase. These include the Genomic Data Commons, the Proteomic Data Commons, and The Cancer Imaging Archive.
Landmark MVP study hits quarter-million enrollment mark

Male breast cancer: A rare disease, on the rise

International study involving VA yields new insight on schizophrenia genes

Chemo without the side effects: Scientists look to gold, toad skin for answers
APOLLO capitalizes on the respective strengths of the three agencies in science and health care. NCI has cutting-edge expertise in the complex data-handling and analysis involved in proteogenomics, a relatively new field. DoD has a well-established pathology network for receiving and analyzing tissue samples—plus facilities for DNA and RNA sequencing, and various platforms for analyzing proteins. VA offers clinical research expertise and infrastructure plus a large pool of patients with cancer.
APOLLO is starting operations at 10 military hospitals and one VA site (Palo Alto), with additional VA sites likely to be on board by late 2019. There is also one civilian hospital involved, Anne Arundel Medical Center in Maryland. Cancer patients at these facilities can agree to have their information—including molecular results from their tumors—added to the growing APOLLO research database. All information is coded, so patients are not personally identifiable to researchers. The enrollment target is 8,000 patients over five years.
The effort will work hand in hand with VA’s Precision Oncology Program. Through POP, increasing numbers of VA patients with cancer are having their tumors genetically analyzed, so their physicians can prescribe more targeted therapies, or so they can be referred to appropriate clinical trials.
POP has its own research arm, called RePOP, supported by the Cooperative Studies Program within the VA Office of Research and Development, and run through the Boston CSP Center. RePOP is an active contributor to APOLLO. At a fall 2018 conference of the American Medical Informatics Association, a RePOP team described how they “successfully moved de-identified clinical, genetic, and imaging data for 113 consented [patients]” to the newly created Precision Oncology Data Repository within VA. From there, the data can be shared with other repositories, such as the Genomic Data Commons—created at the University of Chicago and supported by the National Cancer Institute—to support cancer research outside VA.
If there’s an explosion right now in precision oncology research, it’s inspired in part by the real-world advances that have already taken place. Genetics has a firm foothold in routine cancer care. For instance, a cancer drug called irinotecan is given in lower doses to patients with colorectal cancer who test positive for a certain gene variant. Their bodies make less of an enzyme that metabolizes the drug. One recent boost to the field came last year when the Centers for Medicare & Medicaid Services approved coverage of a new type of genomic testing for patients with advanced cancer, so they could be matched with targeted therapies based on their genes.
But the ambitious APOLLO project will go far beyond genes. For starters, there are proteins to consider. Hence the “proteogenomics” part of APOLLO’s name.
Shriver, a cancer surgeon and researcher who is also a retired colonel from the Army Medical Corps and decorated combat Veteran, uses a military analogy to explain the link between genes and proteins, and what it means in cancer.
“If you go back to basic high school biology,” says Shriver, “there are three molecular components of life: DNA, RNA, and protein. DNA is the instruction manual. The proteins are the action officers, the troops. They're out there in the cell doing all the work. And they may or may not listen to the boss [the DNA, or genes].
“Then you have the RNA—they're the messengers, the troops sending the message from the DNA to the proteins. But again, the proteins don't have to listen.”
In other words, cancer involves not only mutations—or abnormal changes—at the gene level, but foul-ups by proteins.
If you imagine genetic analysis being complicated, try adding proteins into the picture. It’s like going from arithmetic to calculus.
There are some 25,000 genes in the human genome, but among them they code for more than 1 million different proteins. Proteins start off as long chains of amino acids. Once they get their marching orders from DNA, via RNA, they need to fold into intricate 3D shapes—unique combinations of coils, fans, zigzags, tubes—that enable them to carry out precise jobs within the cell. All the while, they undergo any number of chemical changes.
The process is delicate and prone to error. “Proteins are finicky,” in Shriver’s words. When things go wrong, cancer, or other diseases, can result.
If APOLLO’s overall design is complex, sorting out what’s going on at the protein level is especially so. The task demands rigorous and consistent methods across all sites, and at all points in a patient’s care. For example, if a Veteran in APOLLO gets a biopsy, there are time constraints that don’t apply in routine cancer care.
“It's a high-end bio-banking effort because these proteomics platforms require very meticulously acquired samples, beyond standard pathology practice,” says Shriver. “You just can't walk into a VA which may have a great pathology team and say, hey, start doing this, because now you've got to worry about getting these samples within 30 minutes from the OR. They've got to be handled differently at the bench. The clinical diagnosis always takes priority and comes first, of course, but then the pathologists have to be comfortable with our expectation that any remaining, unused tissue from consenting patients be processed for research, including APOLLO.”
Genomic and proteomic findings are among hundreds of data points that will go into the APOLLO information base for each patient (see sidebar). Aside from an array of more conventional data—like patient demographics, radiology reports, and drug prescriptions—APOLLO will delve into “things that have never been looked at before” on a large scale, says Shriver.
One example is tumor biophysics. How does a tumor—a mass of cancer cells—behave from a biomechanical standpoint? What factors govern how those cells migrate from one body tissue to another—for example, from breast to bone?
That area is of special interest to Dr. Jerry S.H. Lee, one of APOLLO’s leaders and a chemical engineer by training.
He talks about how different body tissues have different “squishiness.” Cancer cells can move about more fluidly in one type versus another. When a cell exits a solid tumor and enters the blood circulation, it’s like “running onto 495 (the Capital Beltway)—although probably not during rush hour,” he says.
“Why and how does it do that? How much of that is a protein, and how much is destined by genes? How does the cell know, hey, this is my stop?”
Lee hopes APOLLO will unearth answers to these and other questions on the frontiers of cancer science. And he expects that one day, cancers will be classified not by where in the body they take root, but by their unique genetic and overall biochemical signatures. And drugs will be developed and marketed accordingly. “It won’t be, here’s a lung cancer drug, here’s a prostate cancer drug. But rather, here’s a drug that targets this molecular signature,” says Lee.
He points to two recent FDA approvals for drugs targeting genetic biomarkers as evidence of the practicality of this vision. Regarding its 2017 approval for Keytruda, the FDA said it was the first time the agency had “approved a cancer treatment based on a common biomarker rather than the location in the body where the tumor originated.” The second such approval came in 2018, for a drug sold as Vitrakvi. The FDA commented at that time on the emerging trend toward cancer drugs that are “tissue agnostic,” meaning they work regardless of where in the body the cancer exists, as long as a certain biochemical pathway is involved.
Lee, today an associate professor at the University of Southern California and a member of VA’s National Research Advisory Council, previously served more than a decade at NCI, where he helped shape The Cancer Genome Atlas and the Clinical Proteomics Tumor Analysis Consortium. He says lessons learned during those projects will be used to avoid pitfalls in APOLLO.
For example, he says not enough attention was paid to ensuring clean data from the outset. There was too much noise that had to be filtered out later on, through great effort on the part of the Genomic Data Commons. There were often “batch effects”—variations among genetic samples due to differing conditions between one lab or technician and another. Lee was lead author on an August 2018 commentary in the journal Cell that discussed lessons learned from past work and presented best practices for “data harmonization.”
Another obstacle was that computer networking technology was simply not robust enough yet to keep up with the fast pace of the research.
Lee: “At one point in the middle of the program—and this is 2011 and 2012—we were actually FedExing terabyte drives from site to site because that was faster than the bandwidth we had at that time to upload and download the datasets.”
Shriver was also involved in The Cancer Genome Atlas. Like Lee, he learned firsthand about the need to establish optimal data curation from the outset. There’s a different mindset now, he says, among those in the forefront of cancer informatics.
“Our goal in APOLLO is to make sure the data are pristine,” he says.
Another edge of APOLLO over the earlier project is that it will include longitudinal data from patients—for example, pathology samples, radiology images, or other clinical data gathered several times over the course of the illness—rather than data collected only at a single time.
As the repository grows, in all its complexity—multiple layers of molecular results for each patients, plus a plethora of clinical and demographic data—Shriver anticipates that robots and artificial intelligence will play a role in sorting it all out. As of last summer, such technology, through a public-private partnership, had already helped pinpoint the right genetics-based treatment for more than 2,700 Veteran cancer patients.
Vietnam Veteran Clarence Massey is thankful his battle with cancer is long behind him. For others in his cohort who are destined to face a similar challenge, APOLLO and its deep dive into the science of cancer may mean better treatment, and added years of life. For younger men and women—those who served in the most recent deployments, and those yet to enlist—it’s an even stronger bet that APOLLO will yield a payoff, should they face cancer in their lifetime.
In fact, Shriver is optimistic that APOLLO will have real-world impact relatively soon. Asked when APOLLO will produce tangible results to guide the care of Vets or soldiers with cancer, Shriver replies, “It’s too early now. But talk to us in about a year.”
While genes and proteins are the focus of APOLLO, the project is gathering a far greater array of information. The picture for each cancer patient will be something akin to a 1,000-piece jigsaw puzzle. An article in Clinical Pharmacology & Therapeutics lists at least 10 categories of data to be collected for each participant. Each category of data has numerous subcategories within it, and each of those might be collected at multiple time points during the course of a patient’s illness. The APOLLO team say the scope of the data-gathering is largely unprecedented in cancer research. Here’s a rundown:
Disclaimer: The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions or policies of Uniformed Services University of the Health Sciences (USUHS), The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the Department of Defense (DoD) or the Departments of the Army, Navy, or Air Force. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
VA Research Currents archives || Sign up for VA Research updates